Archives
Interestingly interactions between Smad and RhoA the other
Interestingly, interactions between Smad3 and RhoA, the other small GTPase which is highly homologous to RhoB [9], could not be observed suggesting that the Smad3/RhoB interactions require amino Wiskostatin residues that are unique to RhoB. Combined with our previous observation that the RhoA gene, which is equally important with RhoB for the rapid non-genomic effects of TGFβ, is not a transcriptional target of the TGFβ/Smad signaling pathway [13], our finding indicates that the mechanism of TGFβ autoregulation via Rho/Smad interactions may be specific for RhoB and perhaps for other small GTPases that are induced transcriptionally by TGFβ and are able to interact physically with Smad proteins but the latter remains to be shown.
In conclusion, our data provide evidence in support of a novel mechanism of TGFβ/Smad signaling modulation by the small GTPases RhoB. The proposed model is presented schematically in Fig. 8. According to this model, an increase in the intracellular levels of RhoB, either by TGFβ itself or by other stimuli [9,10,13], leads to the formation of RhoB/Smad3 complexes which prevent the phosphorylation of Smad3 by TGFβ RI and its translocation to the nucleus thus blunting the nuclear responses to TGFβ such as the activation of the cell cycle inhibitor p21WAF1/Cip1 gene which is critical for the tumor suppressor program of TGFβ [2,16]. On the other hand, RhoB/Smad3 complexes in the cytoplasm induce non-nuclear responses to TGFβ including actin cytoskeleton reorganization and EMT, processes which are characteristic of cancer cell metastasis, a process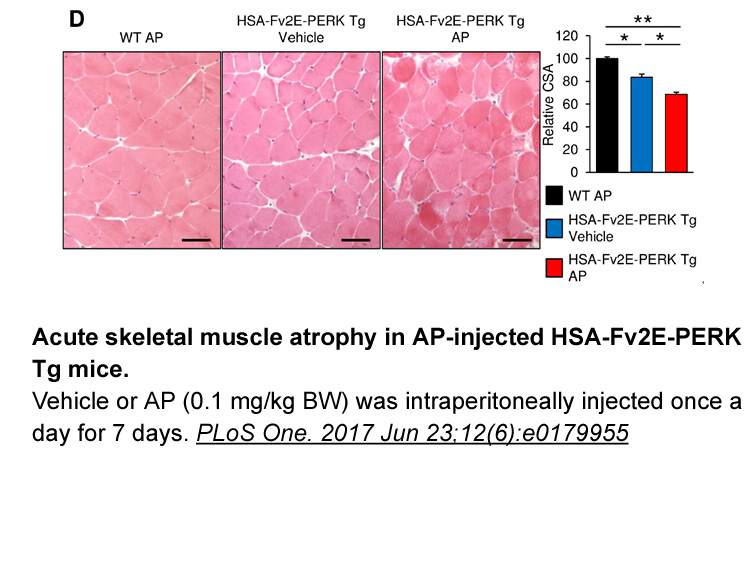 that is known to be induced by TGFβ [27].
that is known to be induced by TGFβ [27].
Main Text
Acknowledgements
Introduction
Paracoccidioidomycosis (PCM) is the predominant systemic mycosis in Latin America with a mortality rate of 25% [1]. Approximately 80% of all the reported cases appear in Brazil, although Colombia, Venezuela, Argentina, Ecuador, and Peru have reported a substantial number of cases [2]. The disease is caused by fungi that belong to the genus Paracoccidioides, and the pathogenic switching of members of this genus is triggered by temperature changes. Once the host has inhaled infective propagules and the fungus reaches the lower respiratory tract, the immune system of the host needs to limit the infection before the disease outcome. Innate immune cells are specialized to produce a set of chemical species, reactive oxygen species (ROS) and reactive nitrogen species (RNS), such as hydrogen peroxide (H2O2), peroxynitrite (ONOO−) and nitric oxide (NO). Thus, the outcome of the infection relies on the ability of the fungus to survive the ROS and RNS produced by the host [3].
It has been demonstrated that NO produced by the host in response to the infection is responsible for controlling fungal proliferation and for avoiding an exacerbated inflammatory response, since the fungal burden in the lungs and liver was higher in mice that do not express inducible nitric oxide synthase (iNOS) compared to wild-type mice [4]. On the other hand, if the host is not able to produce enough RNS to kill the fungus, the host could benefit from the lower levels of RNS produced during disease onset. This fact was demonstrated by the ability of P. brasiliensis to survive and proliferate when exposed to sub-toxic levels of NO, which occurred in  a Ras-dependent manner [5].
Ras GTPases are a family of functionally conserved small proteins that switch between GTP-bound (active form) and GDP-bound (inactive form) conformations. Ras is involved in signal transduction pathways connecting the events from many cell surface receptors to intracellular processes [6]. Depending on the cellular context, Ras activation can stimulate the cell division cycle, morphogenesis, differentiation, or apoptosis [6]. Ras proteins upstream of the cAMP-PKA or MAPK (mitogen-activated protein kinase) pathway are involved in the regulation of filamentation and/or virulence in Cryptococcus neoformans [7] and Candida albicans [8,9], cellular aggregation [10], cell wall integrity [11] and other phenotypes. In P. brasiliensis, two Ras isoforms (Ras1 and Ras2) were characterized with important roles during fungal dimorphism, thermal stress, and in parasite-host interactions [12].
a Ras-dependent manner [5].
Ras GTPases are a family of functionally conserved small proteins that switch between GTP-bound (active form) and GDP-bound (inactive form) conformations. Ras is involved in signal transduction pathways connecting the events from many cell surface receptors to intracellular processes [6]. Depending on the cellular context, Ras activation can stimulate the cell division cycle, morphogenesis, differentiation, or apoptosis [6]. Ras proteins upstream of the cAMP-PKA or MAPK (mitogen-activated protein kinase) pathway are involved in the regulation of filamentation and/or virulence in Cryptococcus neoformans [7] and Candida albicans [8,9], cellular aggregation [10], cell wall integrity [11] and other phenotypes. In P. brasiliensis, two Ras isoforms (Ras1 and Ras2) were characterized with important roles during fungal dimorphism, thermal stress, and in parasite-host interactions [12].